Updating the Set of Optimizing Variables
Besides evaluating the fit between the calculated results and the experimental data, you should also ensure that you have the right number of optimizing variables. If you have too many or too few optimizing variables, then you may not be able to use your model to accurately extrapolate thermodynamic properties in a wide range of temperature, pressure and composition conditions.
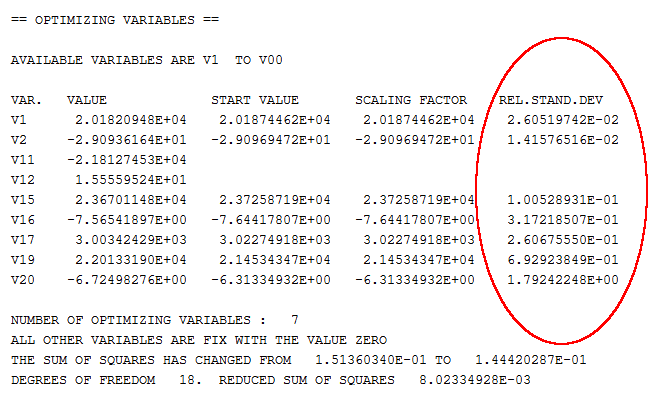
With more optimization parameters, the sum of errors usually decrease, but the parameters also tend to become less precisely determined. A measure of this precision is the Relative Standard Deviation (RSD) of each optimizing variable. However, the RSD is only significant if you have used RESCALE_VARIABLES followed by an optimization which converges and the values of the variables do not change much. The RSD tells you how much the parameter can be changed in either a positive or in a negative direction without changing the reduced sum of errors with more than one unit.
A large RSD value thus means that the parameter has not been determined well.
If such a significant RSD has a value larger than 1, then this typically means that you are using too many optimizing variables. However, the RSD values also depend on the weighting of the experiments, so it is sometimes possible to reduce the RSD by changing weights.
The RSD should not be too low either. If one or more of them is very close to 0, then this suggests that you are trying to optimize the system with too few optimizing variables. You can add optimizing variables interactively using ENTER_PARAMETER in GIBBS. However, it is recommended that you add a sufficient number of variables in the setup file from the start so that do not need to do this. You can fix variables to a value of 0 until you discover that they are needed.
Besides the RSD, you should also keep an eye on the variable values themselves (shown in the VALUE column, next to the leftmost column in the table shown in the preceding screenshot). If the value of a variable is in the order of 1E5 or higher and this variable has been put in the temperature-independent part of an interaction parameter’s Gibbs energy expression, then you may not have the right weights on your experiments or you are using too many optimizing variables. This is also typically the case if a variable with a value in the order of 10 or more (per mole atoms) is in the temperature-dependent part of the Gibbs energy expression of an interaction parameter. Such a high parameter value may lead to inverted miscibility gaps or re-stabilisation of a phase at high temperature. It is important to keep an eye on this since the temperature-independent and the temperature-dependent interaction parameter often varies together. If the temperature-dependent interaction parameter cannot be reliably determined, you may have to fix its value.
Note that even if you have found a good enough fit between calculated and experimental values after an optimization run, a parameter value may suddenly start to change by several orders of magnitude when you use OPTIMIZE_VARIABLES is used again. If this happens, then careful reconsideration of the weighting of the experiments of the set of optimizing variables is required. However, when the optimization has converged and the parameter values remain stable after repeated use of OPTIMZE_VARIALBES, this should usually not happen.